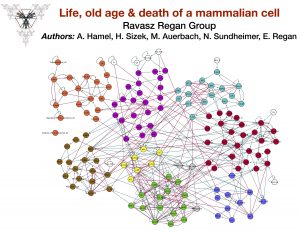
- Boolean Modeling projects in mammalian cells. Building on each other’s IS projects, a subset of my students model life, death, damage, epithelial to mesenchymal transition, and other interesting behaviors of mammalian cells (dots mark the modules a student developed, extended or analyzed):
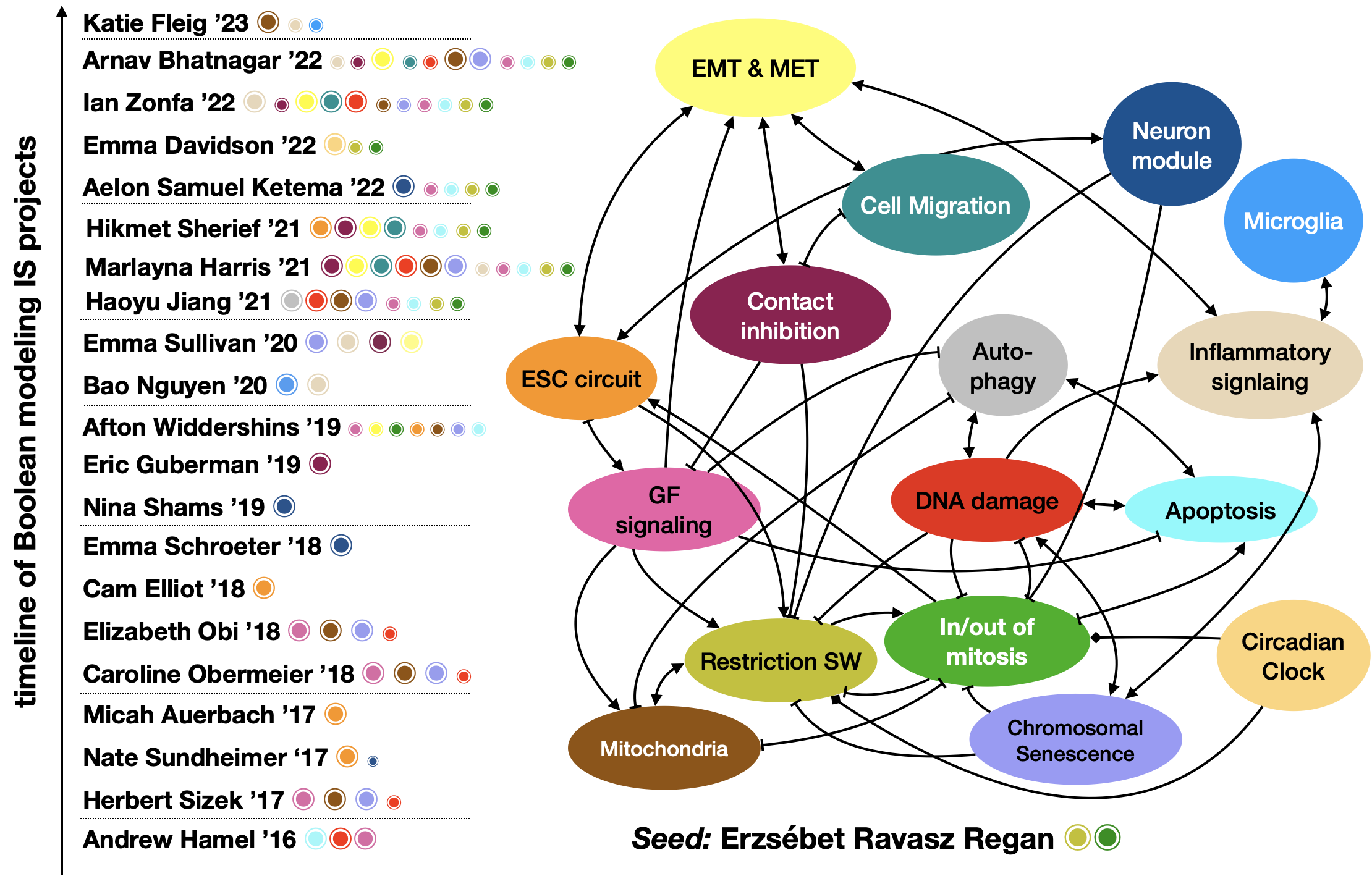
Module-level view with students who contributed to each module during their IS.
- Boolean Modeling in baker’s yeast. During the summer of 2o2o we embarked on a new project to model some of the conserved single-cell functions such as programmed cell death, or apoptosis, in yeast cells. This involved modules related to mitochondrial dynamics, MOMP, and DNA fragmentation, followed by a modular cell cycle model. Stay tuned for its growth as more students take up the challenge.
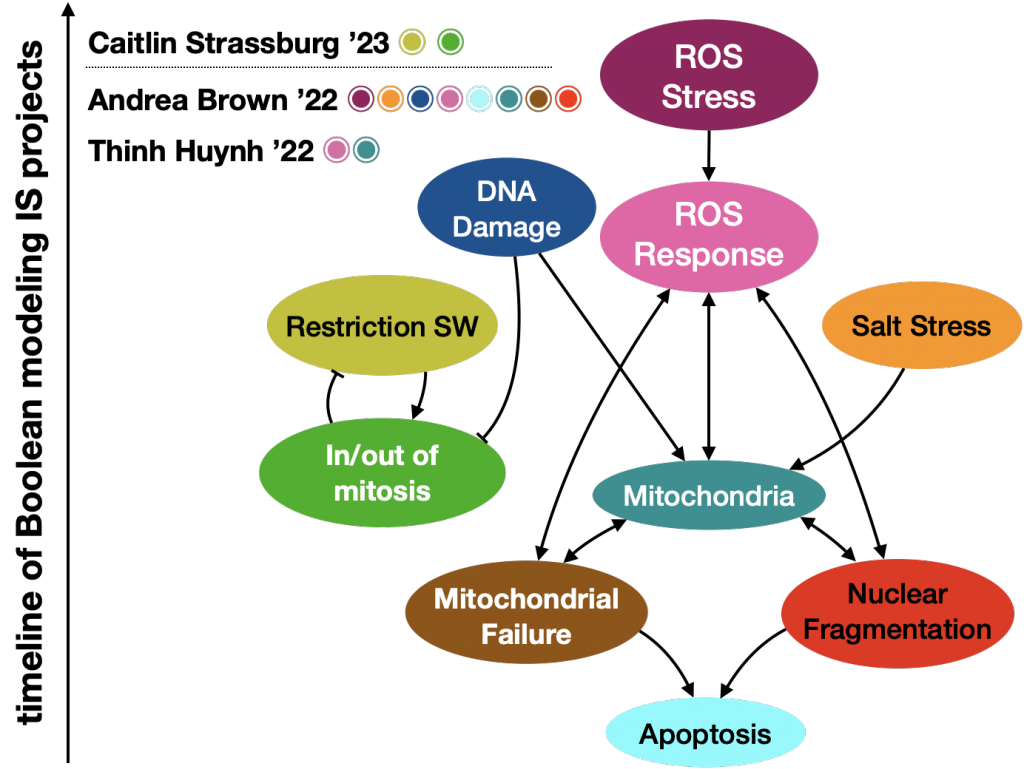
Module-level view with students who contributed to each module during their IS.
- Other IS projects:
- Epigenetics: modeling the dynamics of epigenetic marks at the Oct4 promoter
- Viral communication: modeling the benefits of molecular communication between viruses as they influence the lysis-lysogeny switch of their neighbors
- OARDC projects co-advised by faculty at the OARDC.
- Bioinformatics projects: identification of transcriptional switches from single-cell time series data during development, mapping the functional impact of Phytophthora sojae effector proteins (with W. Morgan, Wooster)
- Publications with IS students:
- Herbert Sizek, Dávid Deritei, Katie Fleig, Marlayna Harris, Peter L. Regan, Kimberly Glass, Ravasz Regan E*, Unlocking Mitochondrial Dysfunction-Associated Senescence (MiDAS) with NAD+ – a Boolean Model of Mitochondrial Dynamics and Cell Cycle Control, 2024 (under review; invitation to Translational Oncology special issue).
- Emmalee Sullivan, Marlayna Harris, Arnav Bhatnagar, Eric Guberman, Ian Zonfa, E. Ravasz Regan, Boolean modeling of mechanosensitive epithelial to mesenchymal transition and its reversal, iScience 26(4), 2023.
- Eric Guberman, Hikmet Sherief, E. Ravasz Regan, Boolean model of anchorage dependence and contact inhibition points to coordinated inhibition but semi-independent induction of proliferation and migration, Computational and Structural Biotechnology Journal, 18: 2145, 2020.
- Herbert Sizek, Andew Hamel, D. Deritei, S. Campbell, E. Ravasz Regan, Boolean model of growth signaling, cell cycle and apoptosis predicts the molecular mechanism of aberrant cell cycle progression driven by hyperactive PI3K. PLoS Computational Biology 15(3): e1006402, 2019.
- Andrea Wade, CH Lin, C Kurkul, E Ravasz Regan, RM Johnson RM, Combined toxicity of insecticides and fungicides applied to California almond orchards to honey bee larvae and adults. Insects, 10(1), 20, 2019.
2024
- Fareeda Abu-Juam (BCMB) – Probing The Relationship Between Partial Epithelial-to-Mesenchymal-Transition and Cell Shape in Epithelial Cells
- Grant Greene (BCMB) – Boolean Modeling of the Molecular Dynamics Governing the Epithelial and Endothelial Hypoxic Response
- Rue Ressler (BCMB) –
- Nina Reddy (Neurobiology) – Boolean Modeling and Microglial Matters: Replicating Microglial Behavior in Inflammation and Testing Model Validity
2023
- Caitlin Strassburg (BCMB / Mathematics) – Conservation of Dynamic Modularity in the Regulation of Cell Cycle and Apoptosis control in Saccharomyces cerevisiae
- Katie Fleig (Neurobiology) – An Overworked Powerhouse: a Boolean Model of Mitochondrial Dysfunction Associated Senescence in the Context of Aging Microglia
2022
- Andrea Brown (BCMB) – The Seven Deadly Pathways: A Boolean Model of Yeast Apoptosis in Response to Reactive Oxygen Species, DNA damage, and Salt
- Emma Davidson (Neurobiology) – Time Flies: An analysis of Circadian impact upon cell cycle regulation using Boolean modeling and a developmental Drosophila model
- Aelon Ketema Samuel (Neurobiology) – A Knock-out Experiment on a Neuronal Boolean Model
- Ian Zonfa (BCMB) – Boolean Modeling of the Interactions Between Inflammation, Hypoxia, and the Epithelial-Mesenchymal Transition (EMT)
- Thinh Huynh (BCMB) – Characterization of a Potential Novel Redox Partner in S. Cerevisiae Mitochondrial Disulfide Relay System and in silico Elucidating Its Essentiality
- Arnav Bhatnagar (BCMB) – Boolean Model of Mesenchymal Cell’s Resistance to Chromosomal Senescence

 2021
2021
- Hikmet Sherief (BCMB) – Modeling mechano-sensitive differentiation and reprogramming of embryonic stem cells
- Marlayna Harris (BCMB) – Modeling the relationship between epithelial to mesenchymal transition and cellular senescence
- Stuart Ball (BCMB) – Quantifying epithelial tissue fitness with a two-scale agent-based / Boolean model
- Brieanna Jarell & Alyssa Ramirez (BCMB ; joint project co-advised by Stephanie Strand) – Experimental and computational study of the flocking phenotype of yeast and its parallels with EMT
- Haoyu Jiang (Biology) – Investigation of Autophagy: Modeling the Cellular Responds to the Undernutrition Condition
- Paige Sogandares (Biology) – Meta-Analysis of Antibiotic Resistance of Helicobacter pylori in Developing Countries (co-advised by the University of Arkansas for Medical Sciences)
2020
- Bao Nguyen (Neuro-BIO) – Modeling neuroinflammation by microglial activation, resolution of inflammation and return to a resting state
- VIDEO summary for non-experts (in Vietnamese with subtitles & live action!)
- Emma Sullivan (Biology) – The senescence associated secretory phenotype promotes epithelial mesenchymal transition in neighboring cells
- Qaiser Zaidi (BCMB) – Computational prediction of developmental switches from single-cell time-series RNA-seq data
- Vi Huynh (BCMB & Math) – Computational Analysis of PsAvh172 Data to Investigate the Targets of Phytophthora sojae in Soybean Host — A Hierarchical Clustering Evaluation of Yeast RNA-Seq Data
2019
- Afton Widdershins (BCMB) – Investigating the Development and Behavior of Heterogeneous Tumors
- Carolina Shams (Neuro-BCMB) – Defying the System: A Boolean Model of Neuronal Fate Decision and Aberrant Cell Cycle Re-entry in Alzheimer’s Disease
- Eric Guberman (BCMB) – A Boolean Model of Contact Inhibition of Proliferation
Afton and Eric presenting their IS work at NetSci’19 in Burlington, Vermont
2018
- Caroline Obermeier (Biology) – Modeling the Effect of p16 on Two Types of Senescence
- Elizabeth Obi (BCMB) – Modeling the Effects of p14 and p53 on Common Oncogenic Pathways
- Emma Schroeter (Neuroscience) – A Boolean Model of Neuronal Death upon Cell Cycle Re-entry
- Campbell Elliott (BCMB) – Using a Boolean Model to Identify Requirements for Successful Yamanaka Reprogramming
- Brendan Kelley-Bukovac (Biology) – Should You Lyse your Neighbors’ Houses? This Model Says ‘No’: An In Silico Examination of the Interactions between Bacterial and Viral Populations in the Presence of the Arbitrium System for Viral Communications

 Left: Emma, Caroline and Cam presenting their IS work at Recomb’18 (Paris); Right: Elizabeth, Emma and Caroline after an amazing joint IS Symposium talk where they tied their very different IS projects into a single story.
Left: Emma, Caroline and Cam presenting their IS work at Recomb’18 (Paris); Right: Elizabeth, Emma and Caroline after an amazing joint IS Symposium talk where they tied their very different IS projects into a single story.
2017
- Micah Auerbach (BCMB) – A Boolean Model of Early Stem Cell Fate Decisions
- Laura Cremer (Biology) – Modeling the Dynamics of Epigenetic Changes on Key Pluripotency Genes in Response to Reprogramming
- Herbert Sizek (BCMB) – Modeling the Commitment to Senescence Through Mitochondrial Fusion and Chromosomal Restructuring
- Nathan Sundheimer (Neuroscience) – A Boolean Modeling Approach to Understanding Synergies Between Cell Phenotypes in Early Neurogenesis
- Andrea Wade (BCMB) – Insecticide Fungicide Interaction and Synergistic Toxicity in Honey Bees (work @ OARDC, Reed Johnson’s Lab)
2016
- Andrew Hamel (BCMB) – A Dynamical Systems Approach to Understanding Synergies Between Thyroid Cancer Mutations
- Heather Skinner (BCMB) – The Putative Regulatory Role of Hydrogen Cyanide in Ethylene Signaling and Biosynthesis in Arabidopsis seedlings and tomato fruit (work @ OARDC, Christopher Taylor’s Lab)