A Dynamical Systems Approach to Understanding Synergies Between Thyroid Cancer Mutations
Class of 2016
Biography:

- Education
- Boston University, MS in Bioinformatics. Expected graduation: January 2019
- The College of Wooster, BA in Biochemistry and Molecular Biology, 2016
- Professional experience
- experience with Python, R, Bash, SQL
- bioinformatics internship, Massachusetts General Hospital
- Americorps Member, City Year, 2016-17
IS Thesis Abstract:
In malignant cells, mutations leading to excessive proliferation cause genomic instability, which further disrupt the cellular regulatory network. Consequently, this leads to signal transduction pathways that no longer respond to critical environmental signals. In papillary thyroid cancer (PTC), the BRAFV600E mutation stimulates rapid cell growth and proliferation in the absence of growth stimulations, triggering tumorigenesis. Often, BRAFV600E co-occurs with genomic alterations in other signaling pathways, exacerbating abnormal PTC behavior and resulting in unpredictable responses to BRAF therapy. Here, I use a modular Boolean modeling approach to explore the synergy between two frequent thyroid cancer mutations: BRAFV600E and genomic amplification of MCL-1. This model is distinct from previously published cancer signaling models in that it is built with a rigorous definition of modularity in mind. As expected, in the presence of growth factors, healthy cells in our model respond to mild DNA damage with a transient cell cycle arrest. In contrast, BRAFV600E-positive cells proliferate in the absence of growth factors and protect themselves from transient DNA damage by exiting into a quiescent state. This may represent an unrecognized mechanism by which BRAFV600E–positive tumors withstand poor environmental conditions. In the presence of growth factors, genomic amplification of MCL-1 increased resistance of BRAFV600E cells to short-term DNA damage from UV irradiation. Intriguingly, this protection is dependent on which the cell cycle phase the apoptosis-promoting influences arrive. Our results suggest that the synergy between BRAFV600E and genomic amplification of MCL-1 aids in evasion of apoptosis from apoptotic signals that preferentially kill proliferating cells. From this, we can make testable predictions and challenge the boundaries of what we perceive are well understood in biology.
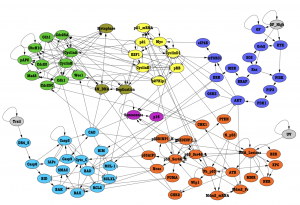 Figure 1.Connected molecular-level Boolean model of healthy thyrocyte. Cell cycle (green, moss, and yellow), cell growth (blue), apoptosis (light blue), DNA- damage (orange), and senescence (purple) switches are linked to create a cellular regulatory system with three inputs (all in gray): GF_High, Trail, and UV. GF_High stimulates cell cycle and cell growth, Trail induces apoptosis, and UV activates the DNA-damage pathway to induce p53-mediated apoptosis.
Figure 1.Connected molecular-level Boolean model of healthy thyrocyte. Cell cycle (green, moss, and yellow), cell growth (blue), apoptosis (light blue), DNA- damage (orange), and senescence (purple) switches are linked to create a cellular regulatory system with three inputs (all in gray): GF_High, Trail, and UV. GF_High stimulates cell cycle and cell growth, Trail induces apoptosis, and UV activates the DNA-damage pathway to induce p53-mediated apoptosis.
Webpage: https://www.linkedin.com/in/andrew-hamel-552996138/
Publications from IS:
- H. Sizek, A. Hamel, D. Deritei, S. Campbell, E. Ravasz Regan, Boolean model of growth signaling, cell cycle and apoptosis predicts the molecular mechanism of aberrant cell cycle progression driven by hyperactive PI3K. PLoS Computational Biology 15(3): e1006402, 2019.
